As scientists and the public struggle with COVID-20 information overload, a Scripps Research team creates an online haven where data is organized, findable and usable.
The volume and speed of COVID-19 discovery around the world is nothing short of astounding. Genetic sequences of coronavirus samples are posted daily to research websites, while thousands of new studies unravel mysteries about the new disease every week.
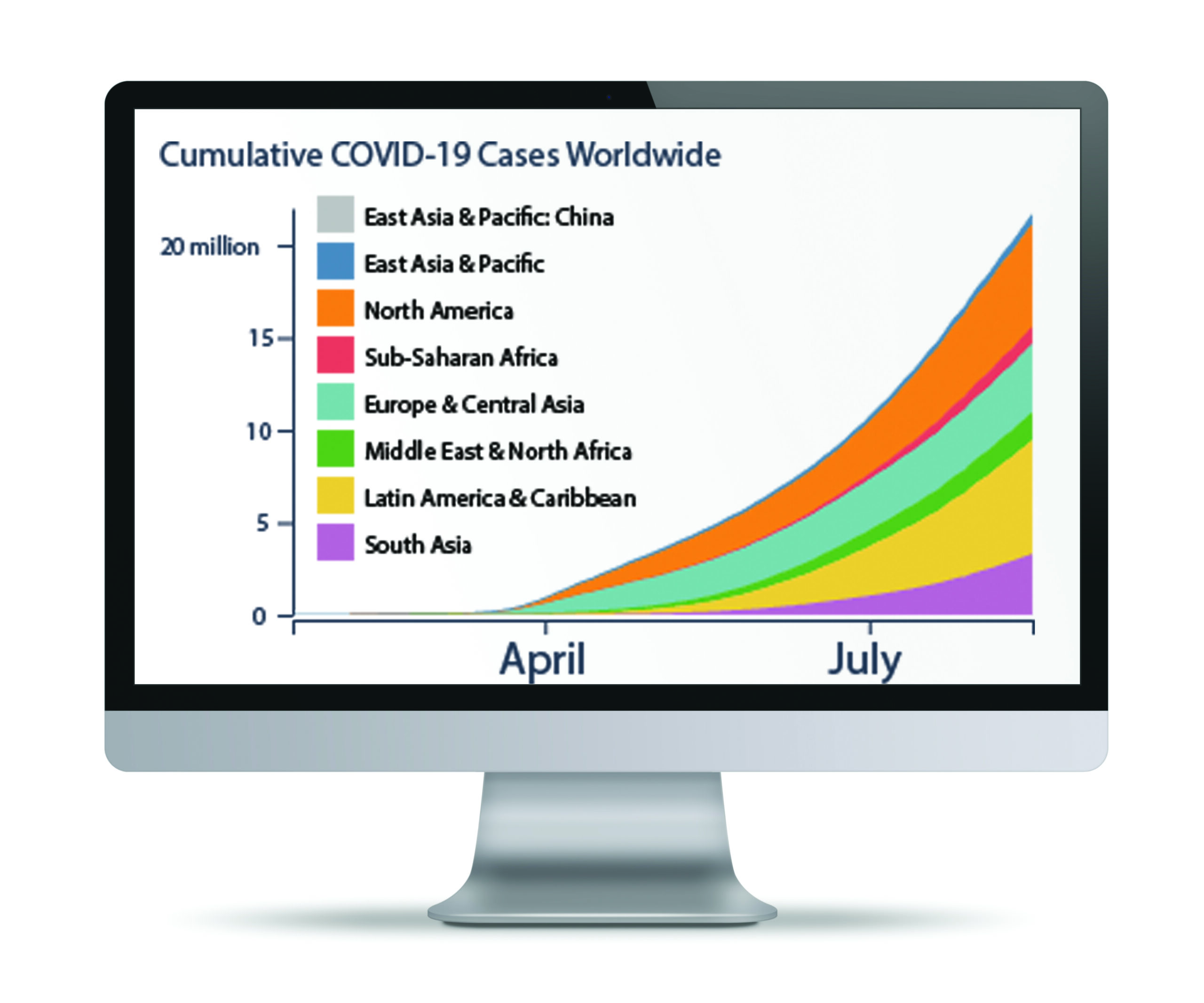
The fact that so much scientific knowledge is being openly shared is a wonderful thing, says Scripps Research data scientist Laura Hughes, PhD. But this “explosion of information” also presents problems—for scientists and the public, she says.
Data is hard to track down from different websites and appears in widely varying formats, making apples-to-apples comparisons an arduous task. For example, a drug might be listed as its marketed name on one website and its generic name on another.
Hughes, who works in the lab of Andrew Su, PhD, collaborated with colleagues on a remedy: a centralized, organized research repository of all things COVID-19. You can find it online at Outbreak.info, which she hopes will become a prized resource for both scientists and the public.
Visually appealing charts and graphs pull information from more than a dozen sources “so it can be understood and accessed faster than before,” Hughes says. “We’re spending a lot of time to make data consistent, which is a very big task. It may sound boring, but it’s necessary to help scientists unite towards a common goal.”
Karthik Gangavarapu, a graduate student in the labs of Su and Kristian Andersen, PhD, and a key member of the Outbreak.info team, says the website will become richer as new information becomes available. The group is creating and integrating computer programs that crawl the web for new data relevant to COVID-19. With help from bots, researchers and citizen scientists can then more easily curate data for the site, he says.
“The website is just one piece of a bigger vision,” Gangavarapu says. “First and foremost, our goal is to help move research forward by making data more accessible. But in the process, we hope that we will help energize and inform the public.”
Calling All Citizen Scientists
“Tapping unused processing power on thousands of unused devises provides us with an incredible amount of computing power to virtually screen millions of chemical compounds.”
—Stefano Forli, PhD
Assistant professor in the Department of Integrative Structural and Computational Biology and director of OpenPandemics project
The value of citizen science—which harnesses the power of the public to gather important research data—is often associated with ecological projects such as the annual Christmas bird count. But the approach is also advancing key health discoveries, especially as scientists seek to understand a completely new and fast-spreading virus. At Scripps Research, here’s how you can get involved.
Use Your Computer to Discover a Drug
Have a computer and an internet connection? That’s all you need to help scientists find compounds that might be effective medicines against COVID-19. The project, called OpenPandemics, is designed and led by Scripps Research and hosted on IBM’s World Community Grid, a computing resource provided at no charge for scientists. Operating unobtrusively in the background of your computer, an app distributes computational assignments via the IBM cloud. Be a part of it: Volunteers need not have any special technical expertise to participate; the process is automatic and secure. Sign up at ibm.org/OpenPandemics.
Detect Outbreaks with your Fitness Tracker
If you use a wrist-worn fitness device such as Fitbit, Apple Watch, Amazfit or Garmin Watch, you can join a study focused on detecting outbreaks of viral illnesses, including COVID-19. By harnessing key data points such as heart rate and activity levels, scientists can improve real-time surveillance. Early detection is critical for effective public health response to infectious disease outbreaks. Be a part of it: Join the study and consent to share your data by downloading the MyDataHelps mobile app. The app “speaks” to your wearable devices to capture relevant data points. Visit detectstudy.org for more information.