Over the last few centuries, the ability to take a closer look into biological processes has gone hand-in-hand with world-changing scientific discoveries. With the invention of the microscope around 1600, scientists, for the first time, accessed an entirely new world of cells, bacteria and other living components.
Today, cutting-edge microscopy and visualization technologies let scientists “see” biological details ranging from the inner workings of viruses to the architecture of protein assemblies. By drawing on the power of computational biology and collaborating across scientific disciplines, researchers are gaining unprecedented insights into the world—as well as what makes us human—like never before.
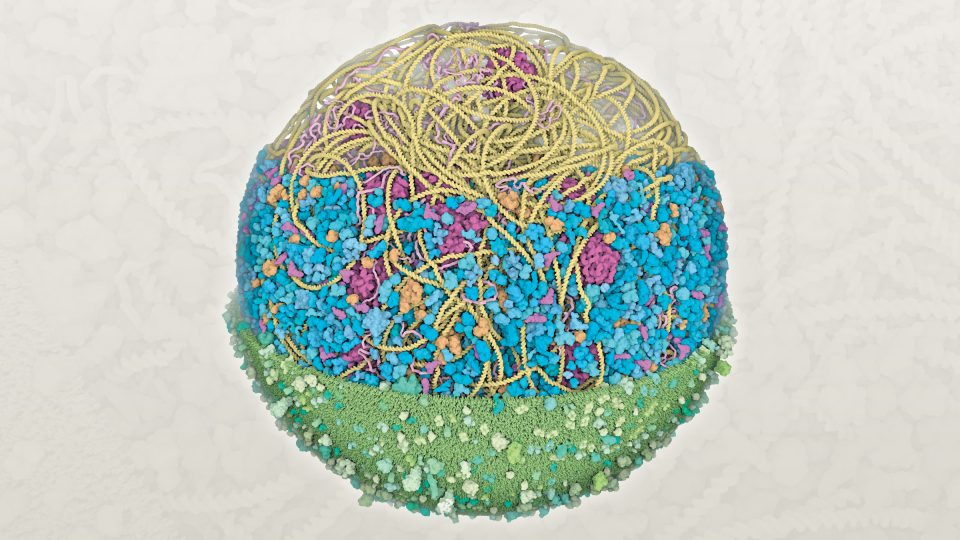
Mycoplasma genitalium
bacterium cell
(radius about 145 nanometers)
Lab:
David S. Goodsell and Arthur J. Olson
Credit:
Martina Maritan, Ludovic Autin, David S. Goodsell, Scripps Research and RCSB Protein Data Bank
Method:
Bioinformatics and computational modeling tools (Mesoscope and CELLPACK)
Description:
Scientists led by Scripps Research faculty David S. Goodsell and Arthur J. Olson created the first 3D structural model of a Mycoplasma genitalium bacterium cell. The upper section shows the protein synthesis machinery, including a long circular DNA strand that encodes the genetic information (yellow), ribosomes that build new proteins (magenta), and many other molecules that assist with the process (orange and pink). The central section includes enzymes (blue) that perform the many chemical reactions needed by the cell. The bottom section shows the cell membrane that protects the cell from the external environment (green), studded with proteins that transport molecules in and out of the cell.
Why it’s important:
Structural models of whole cells integrate experimental results from many different laboratories, including bioinformatics, structural biology and microscopy. Construction of these models has recently become possible by leveraging advanced computational methods from the gaming community. Building on this model, Olson, who holds the Anderson Research Chair, is pioneering 3D computer modeling to similarly model and depict cells in virtual reality. These models help researchers to study biological molecules in their cellular context and allow students and educators to explore the complex environment within a cell.
Learn more:
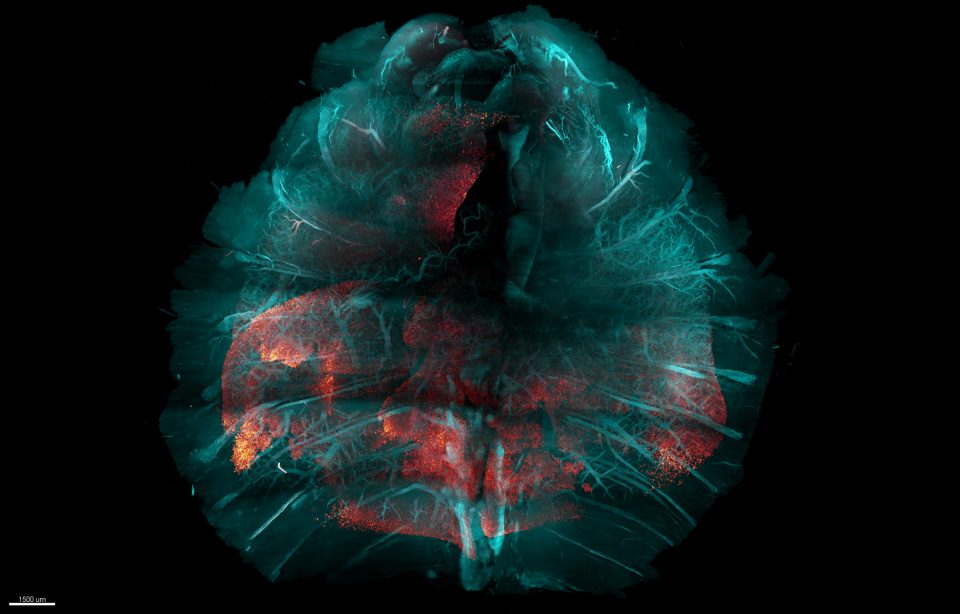
COVID-19 infection in mouse chest
(including all body walls, lungs and internal organs)
Lab:
Li Ye
Credit:
Victoria Nudell and Li Ye
Method:
HYBRiD (hydrogel-based reinforcement of three-Dimensional Imaging of Solvent-Cleared Organs, aka DISCO)
Description:
Li Ye, The Abide-Vividion Chair in Chemistry and Chemical Biology, and his lab have developed an entirely new method for making tissue transparent, which helps researchers to analyze body-wide biological processes, such as infection. This HYBRiD visualization of a whole mouse chest after SARS-CoV-2 infection shows the viral protein (red) spreading through the lungs (red-white scale shows the intensity, with extreme red becoming brighter). Tissue structures like blood vessels and bones (blue) are preserved in this method.
Why it’s important:
Dubbed HYBRiD, this new method combines elements of the two main prior approaches to tissue-clearing technology, and aims to be more practical and scalable than either for large-sample applications. This tissue-clearing technique makes it easier than ever for scientists to render large biological samples transparent to visualize and study healthy and disease-related biological processes occurring across multiple organ systems.
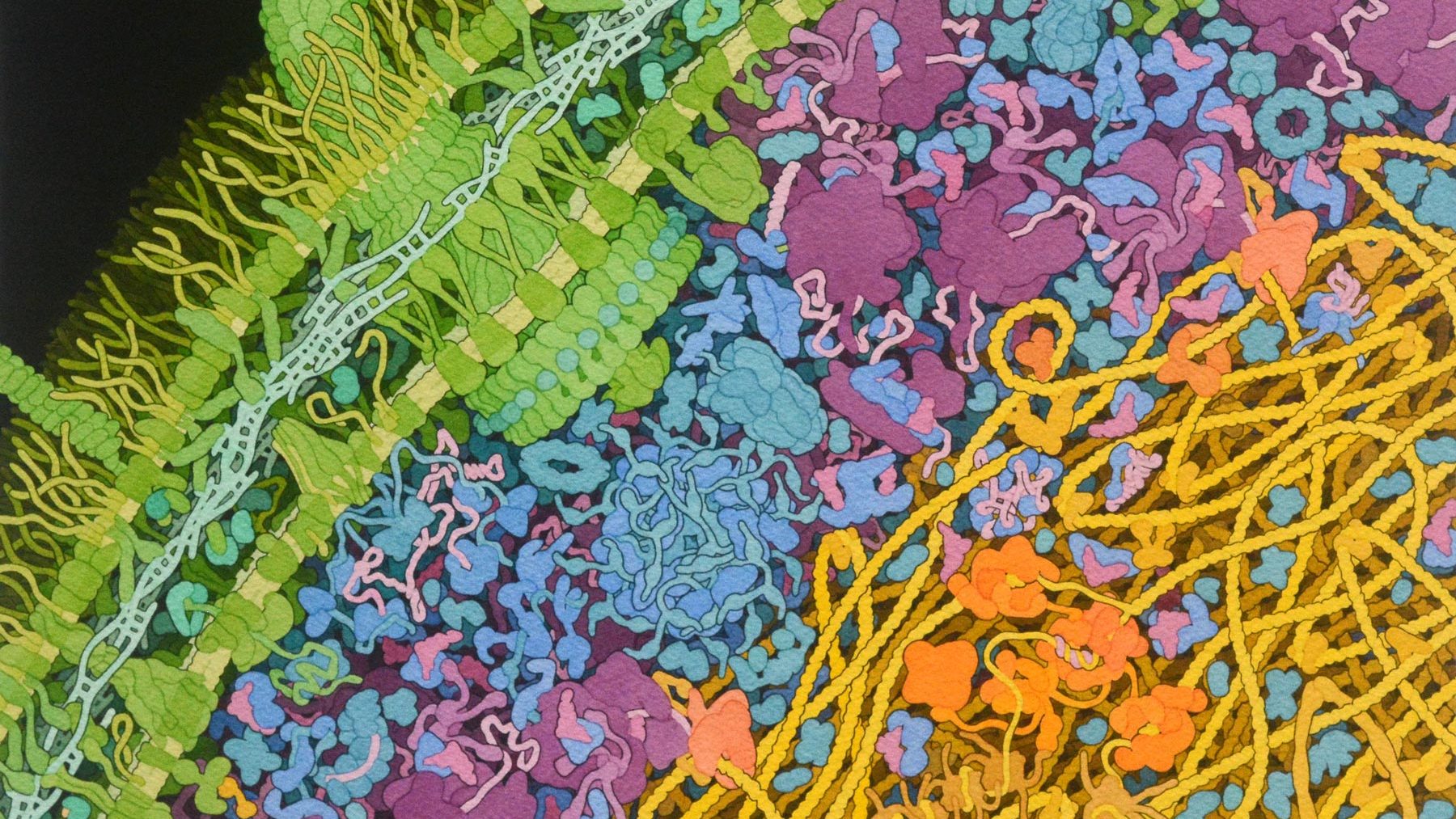
Escherichia coli
bacterium
(225 nanometers wide)
Credit:
David S. Goodsell, RCSB Protein Data Bank
Method:
Hand-drawn watercolor and ink on paper
Description:
This watercolor illustration depicts a cross-section through part of a bacterial cell, showing all the large biomolecules. The cell wall (green) includes a large flagellar motor and many membrane-spanning proteins that transport molecules in and out of the cell. The genome (yellow) interacts with many proteins (tan and orange) that manage the genomic information. Diverse molecules, including enzymes (blue), ribosomes (magenta), and transfer and messenger RNA (pink), perform their biochemical tasks in the space between the cell wall and the genome.
Why it’s important:
Goodsell’s artistic renditions convey the chaotic and crowded nature of a cell. Visualizations such as these provide biologists with a mental model of the cellular context when they conceptualize biochemical experiments or analyze results. They are also widely used for scientific outreach and education to present the complexity of the cell as revealed by structural biology research.
Learn more:
Cryo-electron microscopy (cryo-EM) is a state-of-the-art technology that has ushered in a new era of visualizing the architecture of cells, viruses and proteins to better understand health and disease. By essentially freezing samples and using sophisticated image processing techniques, researchers can reconstruct three-dimensional images of biomolecules at previously unseen resolutions.
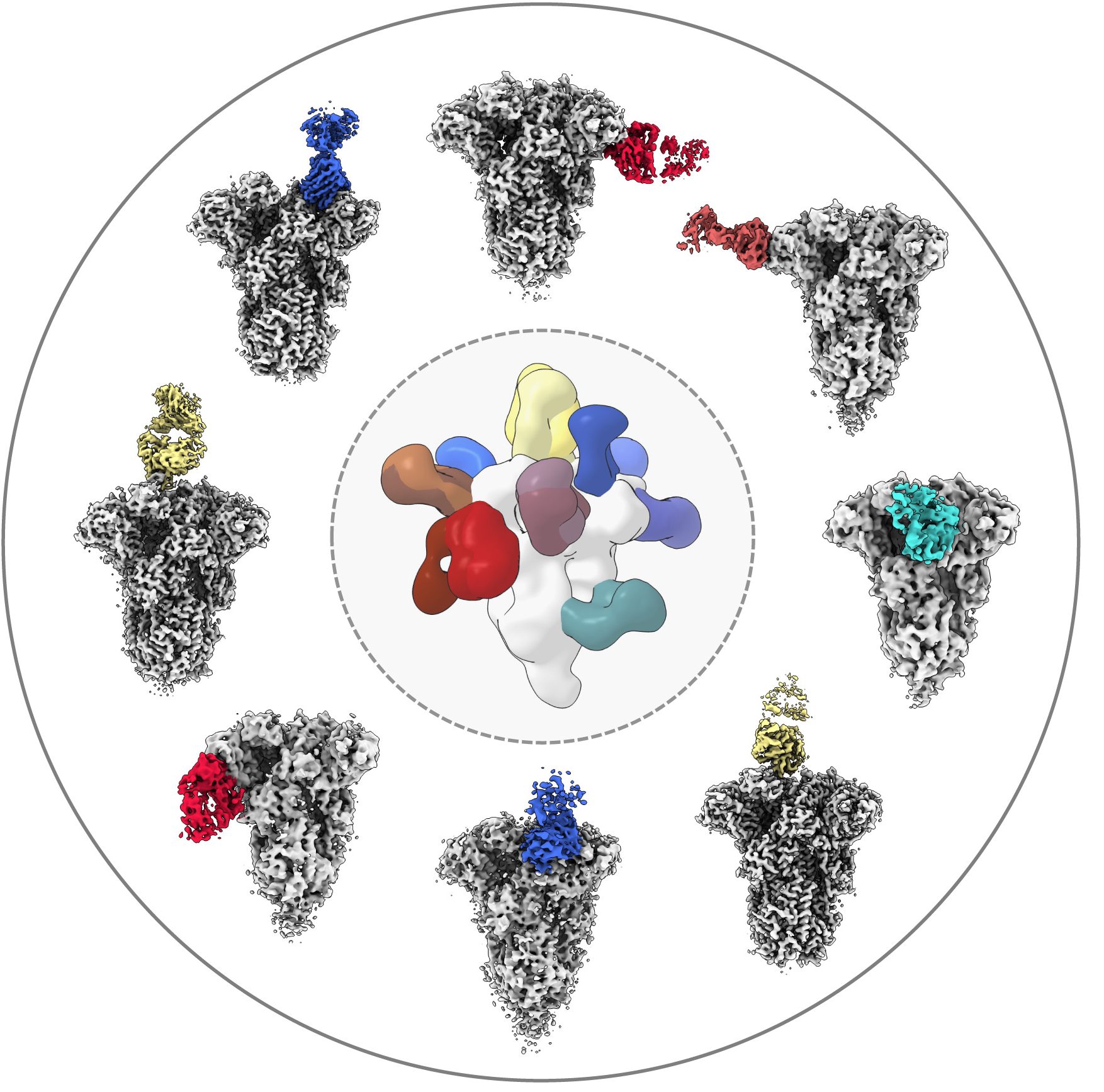
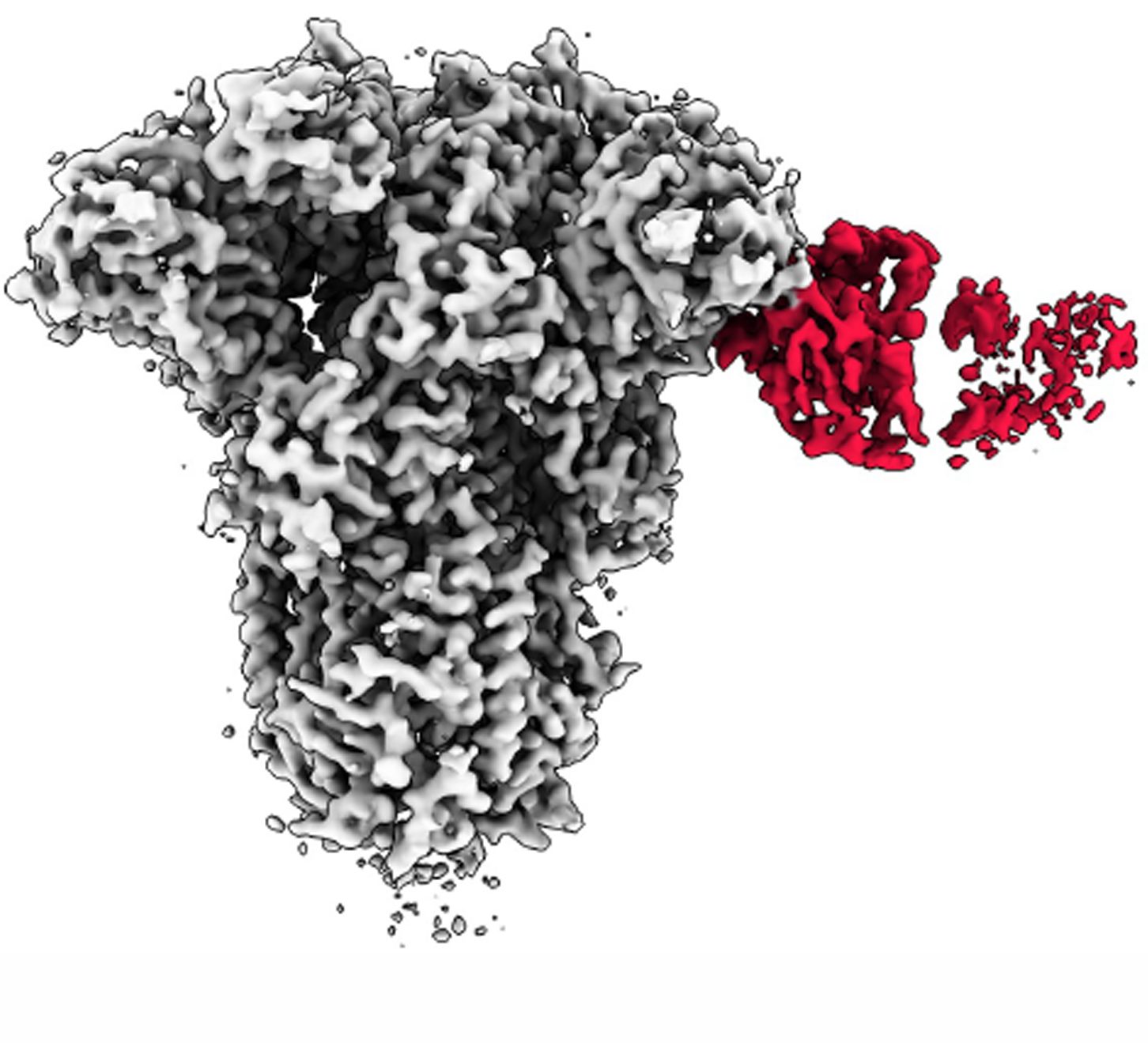
Antibodies and coronavirus spike proteins
(225 nanometers wide)
Lab:
Andrew Ward
Credit:
Andrew Ward lab
Method:
Cryo-EMPEM (cryo-electron microscopy-based polyclonal epitope mapping) and negative-stain electron microscopy
Description:
To analyze how a COVID-19 infection can spur antibodies against common colds, Professor Andrew Ward’s lab used a type of cryo-EM to gain unprecedented visualizations into biological samples at the molecular level. The center image shows a composite of all antibody specificities to the spike protein of the mild coronavirus OC43, derived from multiple donors prior to the COVID-19 pandemic. Some of these samples were imaged by cryo-EMPEM, displayed in the circle. In the circle, the cryo-EMPEM reconstruction of the OC43 spike proteins (grey regions) shows how the spike binds to a specific region of the antibody (colored sections).
Why it’s important:
The team found that COVID-19 patient samples reacted more strongly than the pre-pandemic samples to the spike proteins of OC43. This suggests that a COVID-19 infection might, at least temporarily, boost the number of antibodies you have against common cold-causing coronaviruses and the SARS-CoV-1 and MERS-CoV viruses, all of which are closely related. Gaining a better understanding of how immunity against the broad family of coronaviruses changes with COVID-19 infection can help researchers develop better coronavirus vaccines, both for the current pandemic and for future, related pathogens, according to Ward.
Learn more: